Neuroprosthetics Data Analysis: Project Summary
John Loree
This project was designed to create a rudimentary data analytics software for a Neuroprosthetic controller to be used in my possible 2015-2016 Senior thesis in physics. During this project, I have developed a rudimentary data analysis program which inputs data from an EMG, and calculates two key parameters: peak spike amplitude and spike duration.
Initial Goals
My initial goal for this project was to develop a data analysis program that served the following purposes:
1: Use a Fourier transform to excise the major sources of noise in the signal including movement artifacts and common appliance noise
2: Design a program which can calculate data quasi-continuously (i.e. collect and analyze data in .1 s increments)
3: From the quasi-continuous data find the peak cycle amplitudes and spike duration for each bin and sort the signal into three regimes, a contracting state, a hold state, and a state representing a “rest” position.
4: Ensure the code is easily transferable to other formats / coding languages such that it can be used elsewhere in my thesis.
What I did:
1: The code was successful in sorting and solving for the peak amplitudes and spike duration of the experimental data. However the spike duration is different from the cycle duration (which is more useful) and is not currently calculated. However, the code can easily be adapted to solve for this. The reason it has not yet been modified is that the thresholds and methods by which cycles are delineated are unclear and are part of the larger thesis experiments, therefore they do not fall under the purview of this project.
2: Although the code developed for this project specifically and attached to this submission is unable to run quasi-continuously, the architecture implemented can easily be arranged such that it does run quasi-continuously.
3: The code does successfully sort the data into each of the three regimes, and “tags” them numerically by adding whole numbers to distinguish each of the regimes (0.xxxxx is a contracting state, 1.xxxxx is a hold state, 2.xxxxx is a rest state)
4: The code is easily transferable to other coding languages and formats. With the exception of the Fast Fourier Transform command and language specific syntax, the commands used in the code are language independent. This will allow the code to be transferred to Arduino, which controls the movement of the arm itself streamlining and speeding the execution of each of the experiments.
Methods
The code and parameters for which it solved for were inspired primarily by two papers: Filtering the surface EMG signal: Movement artifact and Baseline noise contamination by Carlo De Luca (2010) and Modulation of in vivo power output during swimming in the African clawed frog by Christopher Richards (2007). The code was written in Matlab, and 3 sample Human EMG data sets were found from a medical database online.
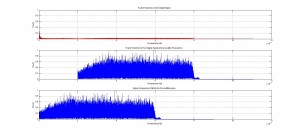
The program has three main sections. First, matrices are created to store the relevant results and initial parameters of the system. Then the input EMG code is chopped into bins .1 seconds in length and each bin progresses through the rest of the code sequentially and independently. From this, the chopped data is passed through a Fourier transform, and the major sources of noise are excised. The excised noise sources are as follows: <20 Hz noise originating primarily from movement artifacts and low frequency pressure waves and 60 Hz noise stemming from most electrical appliances and lights.
The second section solves for the spike amplitudes in each bin. Using the findpeaks command, the locations and magnitudes of each of the spikes are solved for using nested if/else and while loops. The locations of each of the peaks are passed into the third section, while the peak amplitudes are saved as one of the output parameters.
The third section solves for the spike duration and splits the signal into each regime. To do so, a series of nested if/else and while loops are set to proceed from each spike and the beginning of the bin. First, the spike durations are calculated using the locations of the peak amplitudes from the previous section of code. After the spike durations are found, the code cycles back and calculates the regimes where the arm is considered to be either holding or going to a rest state. Any excess time within the bin created as a result of conditions to maintain code stability is then characterized as a hold state, which will allow the physical Neuroprosthetic arm to catch up to the computation in the physical system. The final output durations are then tagged according to which regime they belong into and the results are output in a cycle durations matrix for each bin.
A fourth section, which is only utilized if no spikes are detected in the current bin, goes through the same procedure of finding the hold and rest regime durations for the inactive bins.
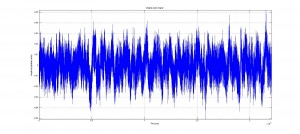
In the code graphs of the original signal, comparisons of the rectified and original signals for each bin and the total program are created to allow the experimenter to have a visual verification of the efficacy of the code.
Results:
As discussed previously, the code was both successful and stable. Following the Fourier noise rectification, the signal was clearer and more legible than the original signal, and the data was still maintained. The peak amplitude outputs are accurate, and are positioned in the output matrices identically to their corresponding spike duration for easy identification and analysis. Similarly, the code successfully found the regime durations in each bin, with hold, contracting and resting states all distinguishable and present in the appropriate locations and placed sequentially.
Analysis of the output signal figure will shows the noise reduction caused as a result of the Fourier noise analysis, and the table, showing the output cycle duration matrix, shows how cycle durations were successfully found. To see larger images, please see either the google drive folder associated with this project, or click on the images themselves.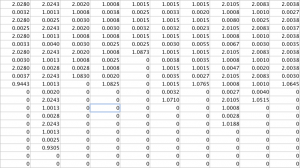
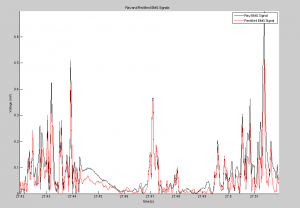
Conclusion and Future Experiments
I am satisfied with this initial foray into Neuroprosthetic data mining. The data outputs were accurate and returned the information that I sought to solve. Furthermore, the architecture of this code is easily transferable to other formats and coding languages, and can be modified to solve for other experimental parameters dependent on the needs of the other experiments.
Although in and of itself, this experiment is extremely successful, and wide number of individual parameters can be calculated by altering the analysis code slightly, the architecture itself may prove unsatisfactory in later experiments. The reason for this is that this code currently only processes data stemming from a single data input. However, there are a number of models for nerve activity that are reliant upon populations and pre-contraction initial conditions (Churchland, 2012), which are significantly more complicated and cannot be solved for using the current architecture. However, I believe that it is not necessary to use that kind of population level model, in which case this architecture is sufficient for later work.
Further modifications to this code will be to change the parameters that the code solves for, as well as transferring the code Arduino such that it can be installed directly into the robot arm. Furthermore, there is a significant amount of information lost through data processing in the name of preventing crashes, which is an inefficiency that I intend to remove in the future. The code developed for this project will serve as an initial, rudimentary data mining code which can be used in a number of different ways elsewhere in my future projects and investigations into neuroprosthetics.
Relevant Files can be found through the link below with headers:
1: EMG_Data_Analysis_427.m
2: SampleEMG1.mat
3: SampleEMG2.mat
4: SampleEMG3.mat
https://drive.google.com/drive/#folders/0B0K4JjJfDy1ffkR4eEZaMzlxb0xvTjRjWDBIUm9iVzZuRmVyeG1aM0RhWHMxcVB6RXZDUFk
References:
1: De Luca, C. Gilmore, D. Kuznetsov, M. Roy, S. Filtering the surface EMG signal: Movement artifact and Baseline noise contamination. Journal of Biomechanics (January 5, 2010) 1573-1579
2: Richards, C. Biewener, A. Modulation of in vivo power output during swimming in the African clawed frog. Journal of Experimental Biology 210 (2007) 3147-3159
3: Lichtwark, G. Wilson, A. A modified Hill muscle model that predicts muscle power output and efficiency during sinusoidal length changes. Journal of Experimental Biology 208 (2005) 2831-2843
4: Computational Physics by Nicholas J. Giordano and Hisao Nakanishi
5: Churchland, M. et. al. Neural population dynamics during reaching Nature (2012) 51-56