The Biology Department held its poster session for senior research this week. Its always exciting to see the wonderful work of our students. Great work, all of you!
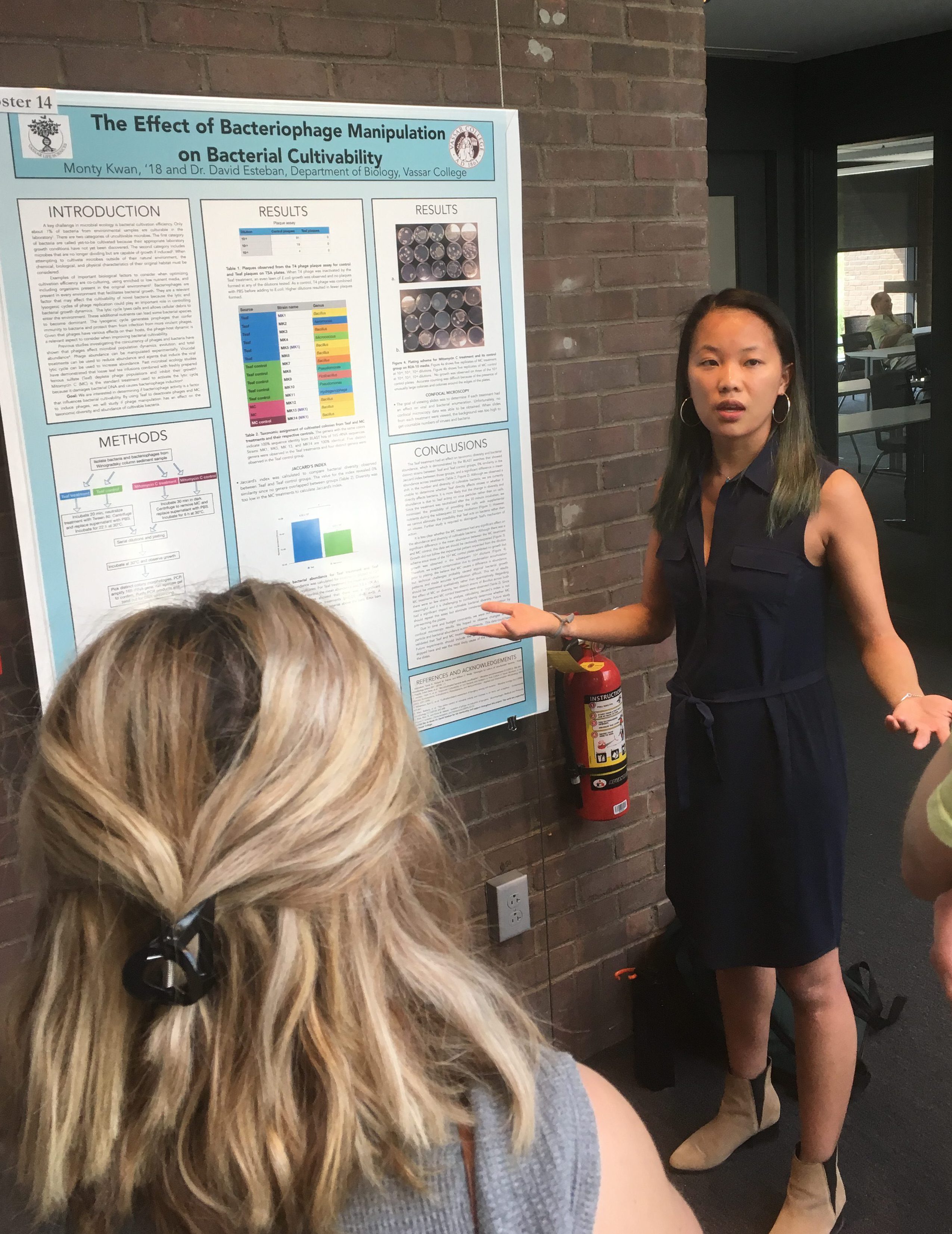
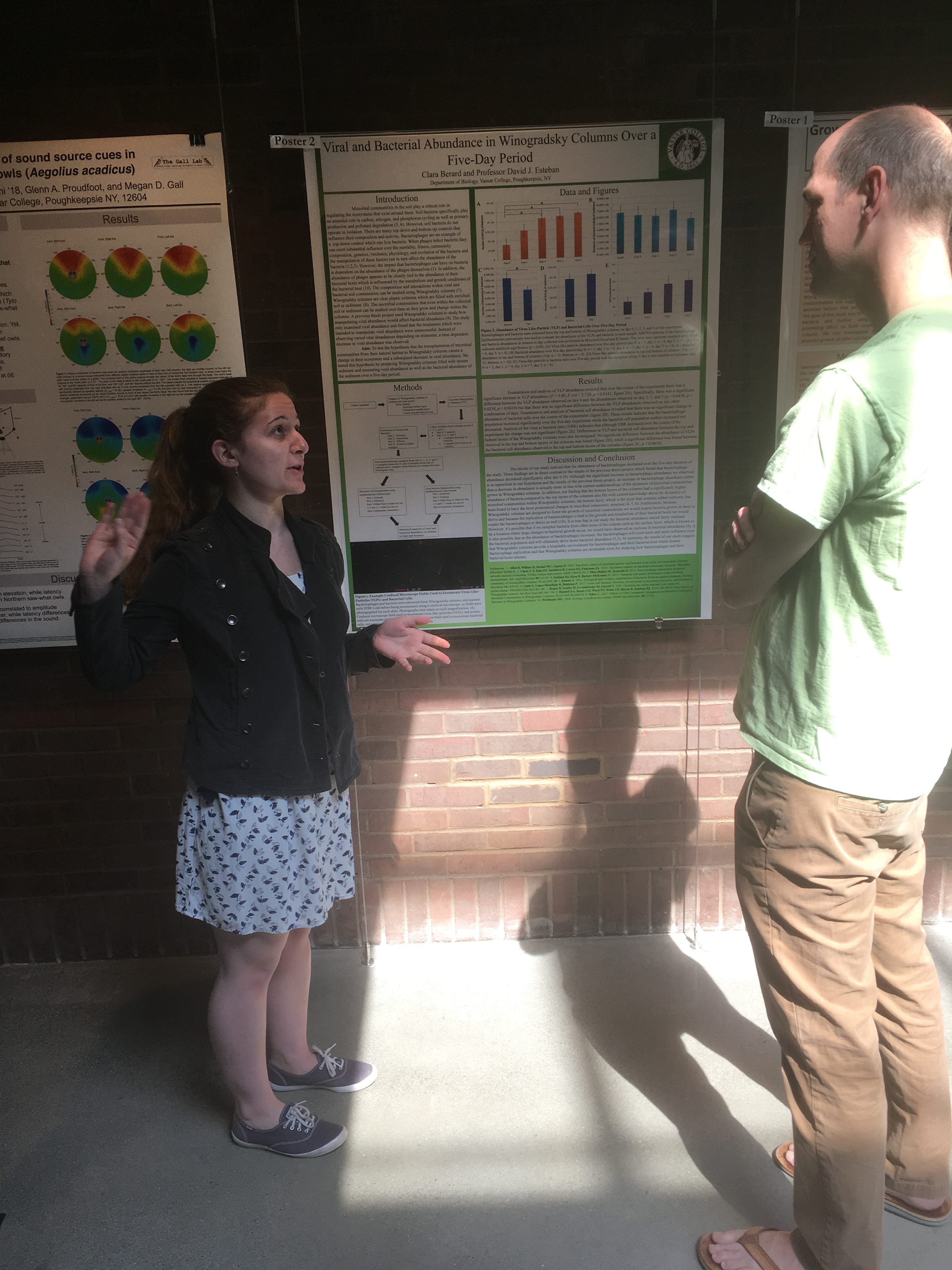
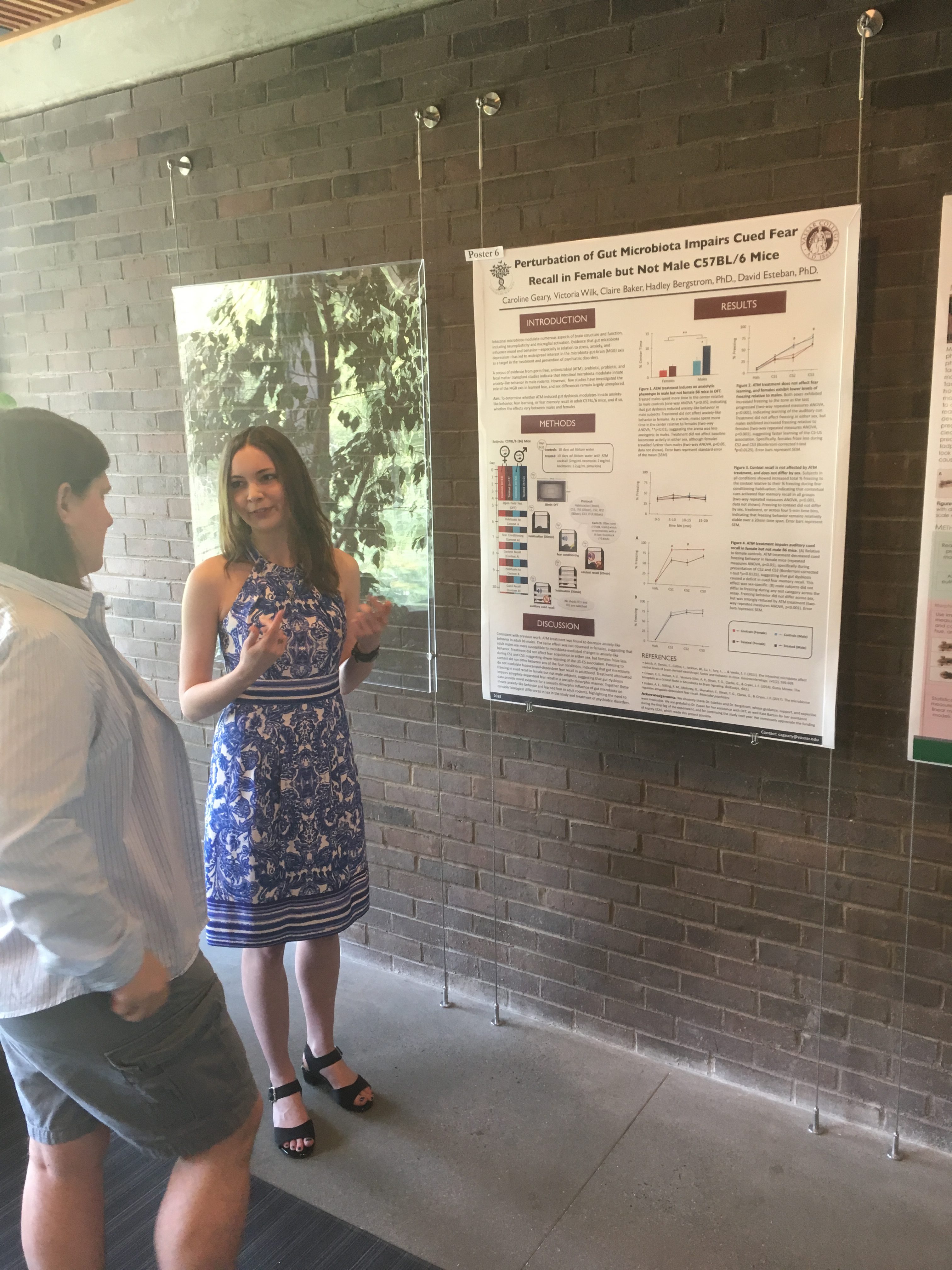
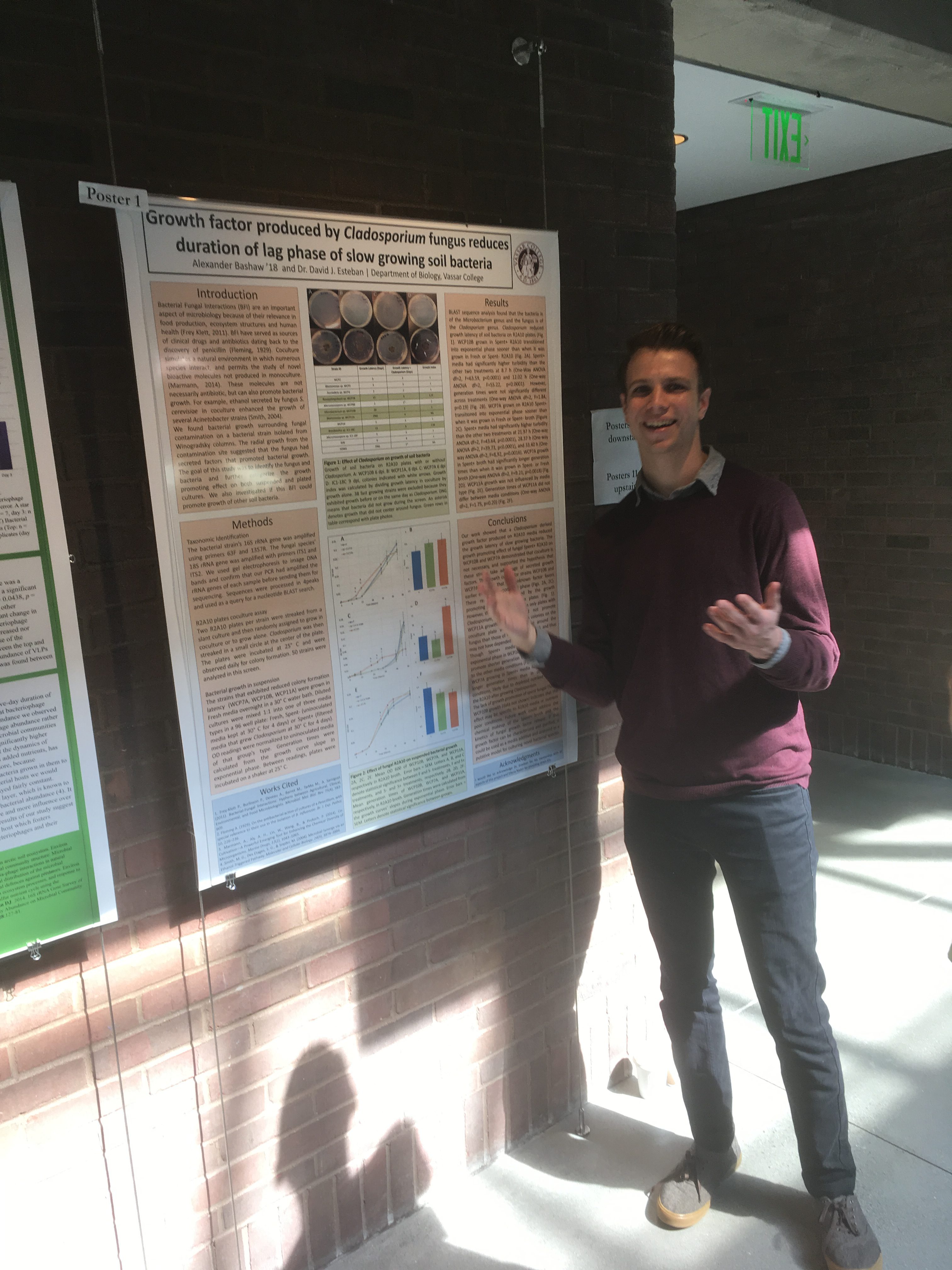
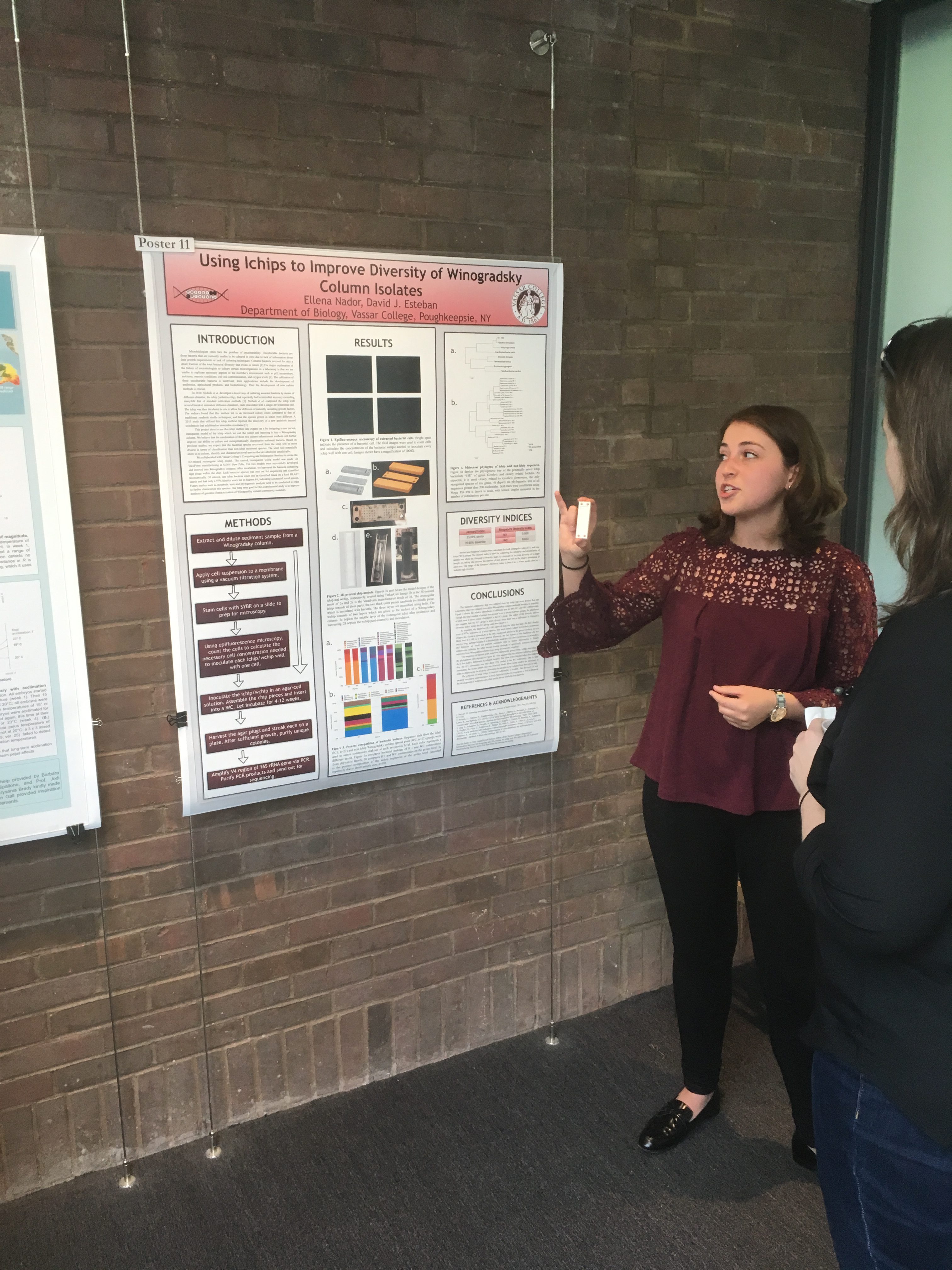
The Biology Department held its poster session for senior research this week. Its always exciting to see the wonderful work of our students. Great work, all of you!





 On April 22 2017 scientists and supporters of science took to the streets in a global rally, the March for Science. Hundreds of thousands marched and participated in events in 600 different cities all over the world to show support for the vital role of science in a democratic and just society and express concern over policies to defund and delegitimize scientific research. Scientists are not typically political activists, so this unprecedented event may signal a shift in the scientific ethos, to one in which scientists see their role extend beyond generating data and new knowledge, but look more to how that knowledge is applied in policy.
On April 22 2017 scientists and supporters of science took to the streets in a global rally, the March for Science. Hundreds of thousands marched and participated in events in 600 different cities all over the world to show support for the vital role of science in a democratic and just society and express concern over policies to defund and delegitimize scientific research. Scientists are not typically political activists, so this unprecedented event may signal a shift in the scientific ethos, to one in which scientists see their role extend beyond generating data and new knowledge, but look more to how that knowledge is applied in policy.
I was honored to lead a team of volunteers in organizing a satellite MfS right here in Poughkeepsie, which drew about 1300 people! The march culminated in a Science Teach-in, at which local researchers, amateur scientists, and organizations mixed with the crowd, discussed topics in science, presented displays, and held activities for folks of all ages.
I will be doing an AMA on Reddit on Wednesday, May 20th at noon EDT. Join the conversation! Ask me anything (well…about microbiology, virology, teaching, research, Vassar, etc…)
http://www.reddit.com/r/IAmA/comments/36mw0z/i_am_david_esteban_researcher_and_professor_of/
 Along with my research intern, Maggie Ginoza, I have launched a crowdfunding campaign to raise funds to sequence the virome of Winogradsky columns, the soil ecosystem model I use to study microbial and viral communities. We’ve gotten a good amount of publicity, check out these links:
Along with my research intern, Maggie Ginoza, I have launched a crowdfunding campaign to raise funds to sequence the virome of Winogradsky columns, the soil ecosystem model I use to study microbial and viral communities. We’ve gotten a good amount of publicity, check out these links:
https://experiment.com/projects/the-dirt-on-viruses-discovering-the-role-of-viruses-in-soil
http://alums.vassar.edu/news/features/2014-2015/150501-going-viral.html
http://miscellanynews.org/2015/04/22/features/internet-a-new-frontier-for-research-funding/
The panel is changing more slowly now, but the oranges and greens are getting richer and the sulfur-enriched mud is showing more and coalescing black spots.
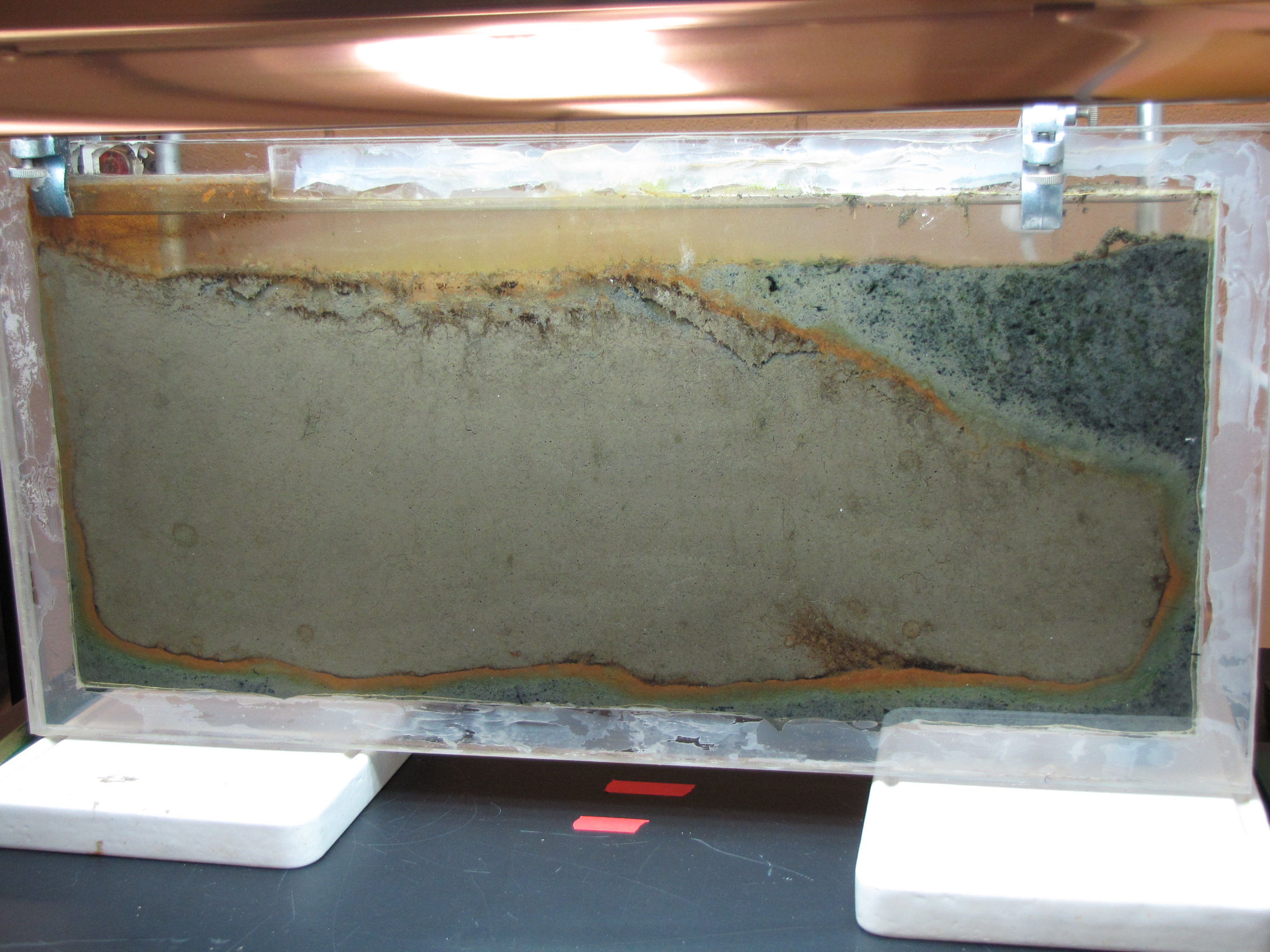 The level of the mud dropped overnight. This could be due to settling of the mud and/or release of gas bubbles. There are some water-filled gaps, and the water level is lower. There is now air above the entire length of the water surface, instead of just at the opening of the panel (top left). Note that there is also a left to right gradient in the water, which was already present before settling. Near the opening, there is brownish/red colonies growing on the walls, probably aerobes (requires oxygen) explaining why its not present further from the opening. The water is murky from left to right, perhaps microbes that can tolerate low levels of oxygen, up to a point near the right side where the water layer is still clear.
The level of the mud dropped overnight. This could be due to settling of the mud and/or release of gas bubbles. There are some water-filled gaps, and the water level is lower. There is now air above the entire length of the water surface, instead of just at the opening of the panel (top left). Note that there is also a left to right gradient in the water, which was already present before settling. Near the opening, there is brownish/red colonies growing on the walls, probably aerobes (requires oxygen) explaining why its not present further from the opening. The water is murky from left to right, perhaps microbes that can tolerate low levels of oxygen, up to a point near the right side where the water layer is still clear.
The Winogradsky panel is developing much more quickly than I had expected. Differences are noticeable from day to day.

The colors at the interface are richer and thicker. The green sulfur bacteria below the orange layer are more obvious now.
 The modified mud layer now has more black spots and there are now some bright green spots, probably green sulfur bacteria. GSB are more tolerant of high H2S concentrations than the purple sulfur bacteria, so are growing well in the areas where the black spots tell us there is H2S. Note the gas bubbles at the bottom center of the image. The squiggly lines are actually paths left behind by the movement of small, red larvae of midge flies, called blood worms. They made their way up to the top, and are now all apparently dead. (And will be food for some bacteria).
The modified mud layer now has more black spots and there are now some bright green spots, probably green sulfur bacteria. GSB are more tolerant of high H2S concentrations than the purple sulfur bacteria, so are growing well in the areas where the black spots tell us there is H2S. Note the gas bubbles at the bottom center of the image. The squiggly lines are actually paths left behind by the movement of small, red larvae of midge flies, called blood worms. They made their way up to the top, and are now all apparently dead. (And will be food for some bacteria).
One of the research projects in my lab is to investigate the microbial communities present in Winogradsky columns. You can read more about what a Winogradsky column is here. Over the next little while, I will be carefully watching the development of the microbial communities over time. This will help me set up a future experiment in which I will take samples over time and determine what bacteria are present and at what abundance to see how the community changes. I will be posting regular updates as the Winogradsky panel develops.
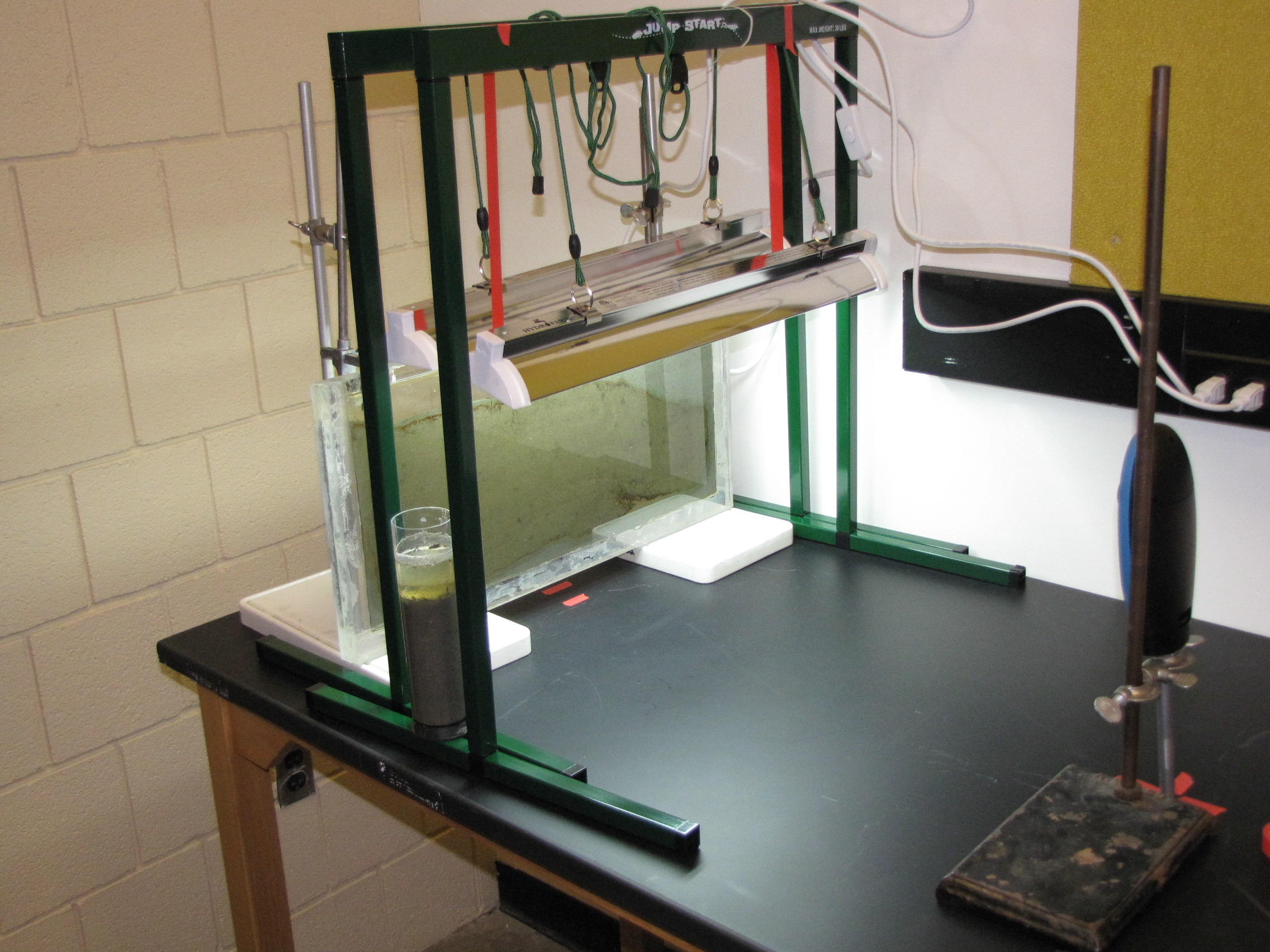
The set-up: the panel is 1′ x 2′ plexiglass with a 1/4″ gap filled with mud from a pond on Vassar campus, grow lamps, and time-lapse camera.

The panel after 2 days. The panel was prepared by adding calcium carbonate, calcium sulfate, and dried leaves to mud from a pond on Vassar campus. We sifted the mud to make a fine slurry. We collected leaves from near the pond and baked them to dry them, then blended in a blender to make as fine a powder as possible. The leaves serve as a source of cellulose. This supplemented mud was poured to a depth of about 4 cm. Then we poured unmodified mud (no additives) on top. Instead of just layering on top though, the bottom layer was displaced, and most of it was pushed to the right side and up to the top. You can see it clearly here: the mud with additives has the lighter grey color.
Note the orange and brownish-orange along the junction between the two muds. This is microbial growth, probably the phototrophic “purple bacteria.” Purple bacteria come in many colors, including red, orange, brown, and yes, purple. They use light for energy but unlike plants they use H2S instead of water (H2O) as an electron source, and produce sulfur instead of oxygen. You can also see it in the little circles where a spot of sulfur-enriched mud got surrounded by unmodified mud.
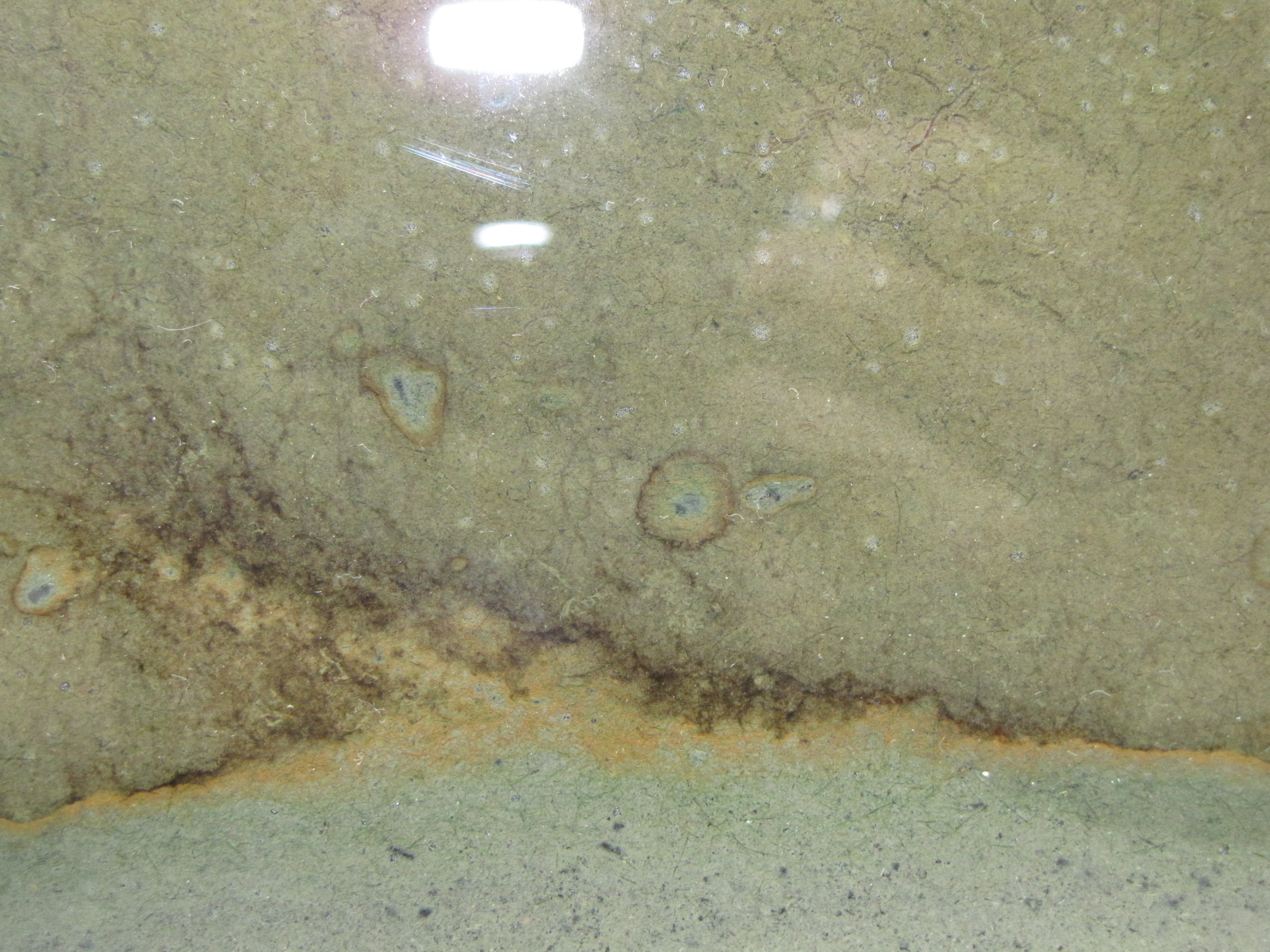 Close-up of the layers. There is a faint hint of green below the orange layer, these are green sulfur bacteria.
Close-up of the layers. There is a faint hint of green below the orange layer, these are green sulfur bacteria.
 Close-up. You can clearly see the layers of color starting to develop. The whitish spots are actually gas bubbles, possibly CO2 or H2 produced from fermentation. That CO2 will serve as a carbon source for autotrophs, and the H2 will serve as an energy source for hydrogenotrophs. Some black specs are showing up in the modified mud. Black is due to microbial production of hydrogen sulfide (H2S), which reacts with iron in the mud producing pyrite, which is black.
Close-up. You can clearly see the layers of color starting to develop. The whitish spots are actually gas bubbles, possibly CO2 or H2 produced from fermentation. That CO2 will serve as a carbon source for autotrophs, and the H2 will serve as an energy source for hydrogenotrophs. Some black specs are showing up in the modified mud. Black is due to microbial production of hydrogen sulfide (H2S), which reacts with iron in the mud producing pyrite, which is black.
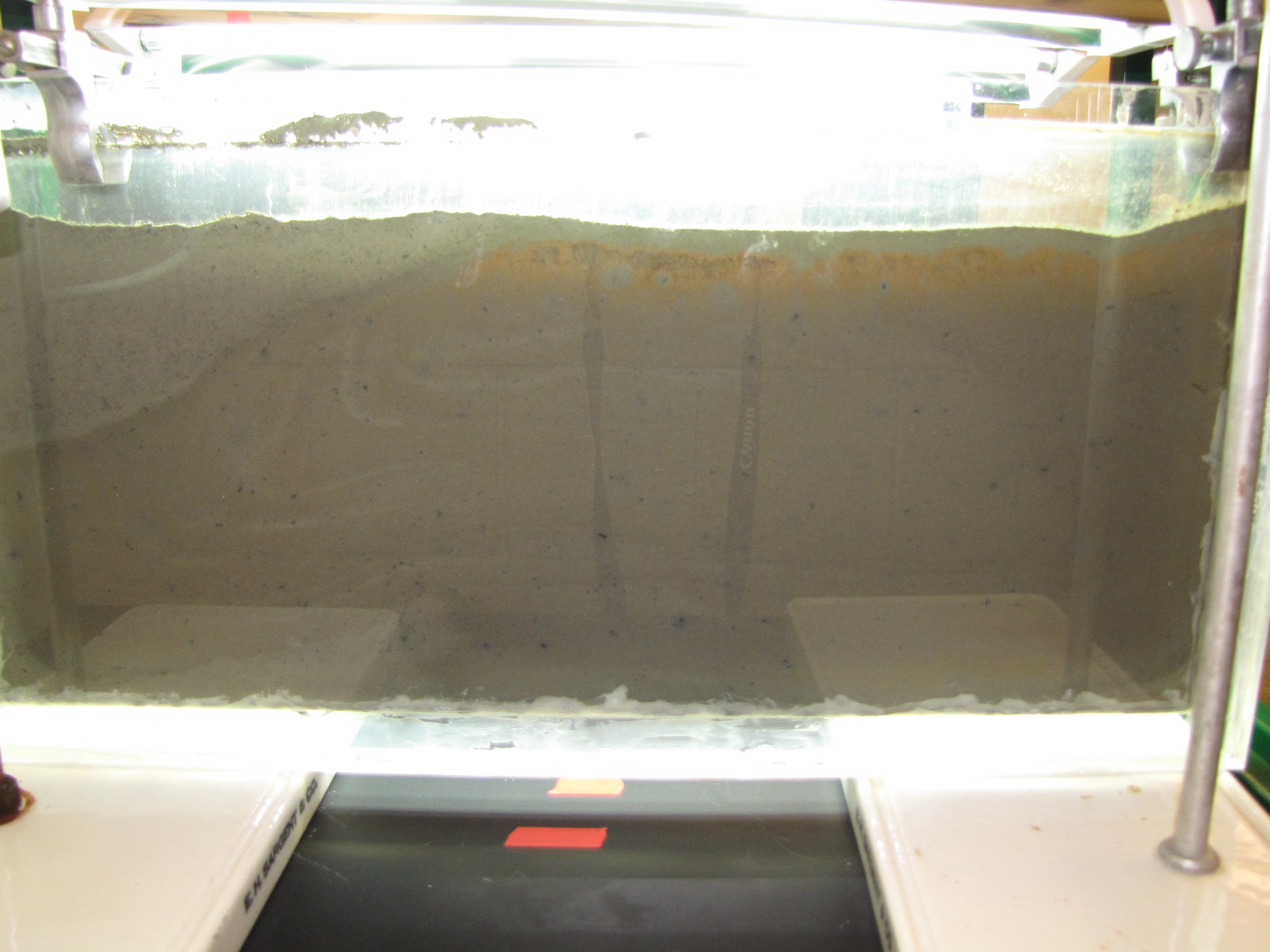 The back side of the panel. This side does not get direct light from the grow lamps. Some gets through on top and you can see the orange colored bacteria at the top. Note that the interface between the muds is less distinct here, showing that the colors on the illuminated side really are phototrophs.
The back side of the panel. This side does not get direct light from the grow lamps. Some gets through on top and you can see the orange colored bacteria at the top. Note that the interface between the muds is less distinct here, showing that the colors on the illuminated side really are phototrophs.
As a bonus question on their final exam, I asked my intro biology students whether or not they would want to have their genome sequenced. The question was:
It is already possible to get your whole genome sequenced and it wont be long before it becomes a routine part of your medical care. Having your genome sequenced can potentially reveal information like increased or decreased susceptibility to specific viral diseases, relative likelihood of developing diseases like Alzheimer’s or Huntington’s disease, or reveal your MHC alleles (which can influence mate choice). Given the opportunity, would you, right now, want to have your genome sequenced? Explain why or why not.
23 students said they would want their genome sequenced, only 6 did not. What I found most interesting is that those in favor and those against often gave the same reasons, it was simply a matter of perspective on whether they viewed it as a positive or negative thing. Here are the common reasons given for wanting to know your genome sequence:
1. Want to know more about family ancestry
2. Finding out about the relative risks of developing certain diseases, so that you can be prepared or do something about it to reduce your risk. In some cases a family history of disease was seen as more reason to know more about your own genes.
3. Curiosity – wound’t it be cool to know?
4. Wanting to give children the best possible chances. Knowing the risk of passing on a genetic disease to children was seen as an important factior in deciding whether or not to have children. Knowing whether a potential mate had “good genes” was also seen as important, or if the combination of genes might be beneficial or detrimental (such as ensuring heterozygosity at the MHC locus). Some saw this as getting an upper hand on natural selection.
Those who did not want to have their genome sequenced:
1. Not wanting to know their relative risk of developing specific diseases. Knowing this could impact the way you live your life in negative ways like worring about diseases you may or may not develop and for which we can do little or nothing to alter its course. A family history of disease was in some cases seen as more reason to not want to know.
2.Do not want genetics to influence importnat decisions like choosing s mate or the decision on whether or not to have children.
3. Privacy and insurance problems. Not everything has been worked out yet with respect to storing or diseminating genomic information, or the laws regarding use of that information by insurance companies. Untill they can be sure that the information will remain private and won’t be used to deny insurance, many thought it safer to wait.
There are obviously many resans for and against genome sequencing, much of it comes down to personal views. However, I think a critical question to consider is what you you really want to know from your genome sequence and will it actually be able to tell you what you want to know? What would do with the information if you had it, what changes or choices might you have to make?
 Slime molds look kind of like vomit but they are pretty awesome. In fact, one of them is affectionately nicknamed “dog vomit slime mold.” While vacationing in British Columbia, my 2 year old son, exploring in the trees and shrubs, exclaimed “cupcake!” Knowing it was highly unlikely that there would be a cupcake there, and not wanting him to eat whatever he found, I rushed over to see a pine cone covered in a thick layer of whitish yellowish stuff. It did, in fact look a little like a piece of rather unappealing cake with thinly smeared white frosting and sprinkles of pine needles and soil from the forest floor.
Slime molds look kind of like vomit but they are pretty awesome. In fact, one of them is affectionately nicknamed “dog vomit slime mold.” While vacationing in British Columbia, my 2 year old son, exploring in the trees and shrubs, exclaimed “cupcake!” Knowing it was highly unlikely that there would be a cupcake there, and not wanting him to eat whatever he found, I rushed over to see a pine cone covered in a thick layer of whitish yellowish stuff. It did, in fact look a little like a piece of rather unappealing cake with thinly smeared white frosting and sprinkles of pine needles and soil from the forest floor.
Looking around, I found several other similar things growing, and some that looked quite different. One looked like coral, another had bubbles of clear, sticky liquid. 
 As I observed them over several days, they changed, all eventually looking alike, and after more days, dried up completely to a pile of dusty powder that easily fell apart when disturbed. These were all the same slime mold, at differnt stages of its lifecycle. I’ve sent these photos to various slime mold experts, and it has been tentatively identified as Brefeldia maxima, the “tapioca slime mold.”
As I observed them over several days, they changed, all eventually looking alike, and after more days, dried up completely to a pile of dusty powder that easily fell apart when disturbed. These were all the same slime mold, at differnt stages of its lifecycle. I’ve sent these photos to various slime mold experts, and it has been tentatively identified as Brefeldia maxima, the “tapioca slime mold.”
Slime molds are indeed slimy, in the picture at the left you can see the slime trail it leaves behind as it moves, but they are not mold. They are actually amoeba. These amoeba can exist in the single celled form but under the right conditions, the single cells will amass together. In some slime mold species, called plasmodial slime molds, the cells will actually fuse together to form a single, giant multinucleated cell. Others, called cellular slime molds, form a multicellular mass. Eventually, the slime mold will differntiate and form fruiting bodies, complex strructures in which spores will develop. Spores can then be released to disseminate the slime mold to new locations. What amazes me is the complexity of this process and the blurring of the lines between unicellular and multicellular organisms. You have a single celled organism, in which many individuals can come together into a multicellular (or single giant cell) form, now behaving as one organism that can differntiate to form complex structures like fruiting bodies and spores. Clearly, we can’t be thinking of microorganisms like these as “simple.”
Some recent studies further demonstrate the complexity of slime molds. The slime mold Physarum was used to map an optimal network between points, in this case the Tokyo metropolitan area. Food sources were laid out corresponding to the communities surrounding Tokyo, and Physarum was placed at Tokyo. The slime mold extended plasmodia, or branches, out to the food sources. At first the branches fan out randomly, making many connections, but eventually most plasmodia disappear, leaving only the most optimal connections between the food sources. The final map of the plasmodia turns out to be quite similar to the Tokyo area metro system. Physarum can also do this with the US interstate highway system, and the Canadian highway network (um, isnt there just one highway in Canada?) So an amoeba can simulate a networks developed by human engineers!
In my lab, a student is exploring using Physarum for a similar purpose. Vassar is looking ahead 50 years or so to plan development of the campus. Can we use the slime mold to provide suggestions to the landscape architects on where to place the pathways between buildings? Can the slime mold help me find the best way across campus? We will keep you posted on what we find!
Contributed by guest blogger: Brooke Schieffer ’12
In September of last year, a group of researchers infected cancer patients with a genetically engineered poxvirus. While this may sound like something out of a horror movie, it is actually quite the opposite: Breitbach et al. were performing a clinical trial to explore new, innovative ways to treat cancer tumors. Some people may be put off by the idea of having live viruses injected into their blood stream, but it’s actually not that uncommon. Indeed, many vaccines are actually live viruses. In fact, the vaccinia virus used in this clinical trial was derived from a vaccine for smallpox.
However, creating an attenuated virus to use as a vaccine and creating a virus that selectively infects and destroys cancer cells are two very different things. Before the scientists can create tumor-killing viruses, they first need to make sure that the virus can infect cancer cells while ignoring normal tissue cells in our body. To do this, they genetically engineered a poxvirus, called JX-594, that could replicate only in cells harboring activation of epidermal growth factor (EGFR)/Ras pathway (many epithelial cancers rely on this pathway). The virus, however, does not lyse the cell as this was only a trial to explore the possibilities of selectively cancer infection, not destruction.
The virus itself was chosen for several different reasons. Firstly, vaccinia (and, consequently, JX-594) is well adapted to intravenous transportation and displays some resistance to antibody neutralization in the blood stream. It can also spread quickly within tissues, making it ideal for infecting tumors (especially solid metastatic tumors). Finally, JX-594 replication is dependent on a commonly activated signaling pathway in epithelial cancers: the EGFR/Ras pathway. Furthermore, to determine if JX-594 was selectively infecting and replicating within cancer cells, the researchers incorporated the lacZ transgene (which encodes β-galactosidase) into the viral genome. They then could track β-galactosidase expression via immunohistochemical staining or tagged antibodies to see where the viruses were replicating in human tissue.
To test the effects of their virus, Breitbach et al. conducted a clinical trial with 23 cancer patients by intravenously injecting them with different concentrations of JX-594. They found their results to be quite promising: in the higher dose groups, they observed selective infection in tumor cells and expression of the β-galactosidase protein. And all with little apparent side effects—the worst of which were symptoms typical of a 24-hour flu. This is the first experiment in which an intravenously injected virus was able to selectively replicate in tumor cells and express a transgene. Of course, this is just the first step on the road to effective treatment. First off, this was only a preliminary study to determine if the virus could selectively infect cancer cells, they did not engineer the virus to kill the cells yet. As of now, it is simply a possible delivery method, not a way to kill cancer cells. But the researchers are hopeful that viruses such as JX-594 will eventually be customizable with proteins or siRNA to treat different types of cancer.
However, there are still many questions to keep in mind moving forward. Will we be able to insert a gene into these viral vectors that only destroys tumor cells? Can this delivery system be modified to infect cancers that do not use the EGFR/Ras pathway? What about the possibility of viral mutations that would allow the virus to infect healthy tissue? And what about the immune system’s role? How will multiple treatments work given the fact that the immune system will build up immunity against the virus because it is invading the body? Even with these questions, this clinical trial was still an innovative and interesting new approach to cancer treatment.
Link:
http://www.nature.com/nature/journal/v477/n7362/full/nature10358.html#/affil-auth
Brooke Schieffer is a senior at Vassar College, majoring in Drama.
Social Widgets powered by AB-WebLog.com.