Science‘s IBI Prize was developed to showcase outstanding materials, usable in a wide range of schools and settings, for teaching introductory science courses at the college level. The materials must be designed to encourage students’ natural curiosity about how the world works, rather than to deliver facts and principles about what scientists have already discovered. With a team of scientists and educators at Carleton College and Vassar College, we developed Web-based Genomics Explorers to help guide students through the process of developing authentic research projects in genomics and bioinformatics. We were awarded the IBI Prize, accompanied by an essay published in the January 25 issue of Science.
All posts by Jodi Schwarz
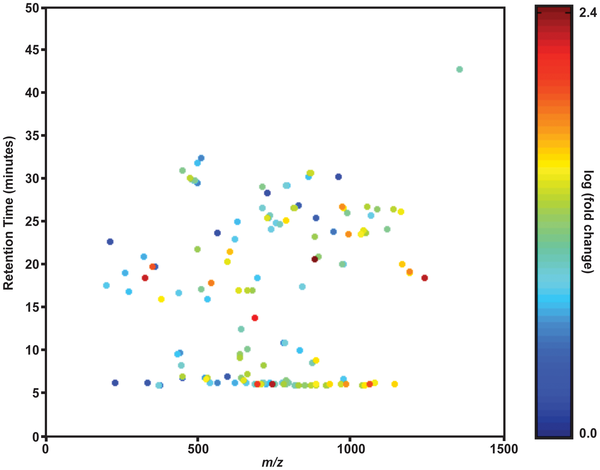
Lipid profiling of symbiosis
Symbiosis in cnidarians is accompanied by significant changes in the lipidome of the anemone host. Congrats to Josh Klein ’12, John Schmeitzel ’12, and Janice Hwang, ’12 for their authorship on this paper just out in PLoS One!
Congrats 2012 Seniors
The Schwarz Lab Senior Thesis students have earned their lab coats! Congratulations!
Integrating Genomics Research throughout the Undergraduate Curriculum
COVER ARTICLE in CBE-LSE: Through a multi-year, collaborative process, we developed, implemented, and peer-reviewed inquiry-based, integrated instructional units (I3Us) adaptable to a range of teaching settings, with a focus on both model and nonmodel systems. Each of the products is built on vetted design principles: 1) they have clear pedagogical objectives; 2) they are integrated with lessons taught in the lecture; 3) they are designed to integrate the learning of science content with learning about the process of science; and 4) they require student reflection and discussion. Eleven I3Us were designed and implemented as multi-week modules within the context of an existing biology course.
Lois M. Banta, Erica J. Crespi, Ross H. Nehm, Jodi A. Schwarz, Susan Singer, Cathryn A. Manduca, and workshop participants.
CBE—Life Sciences Education, Vol. 11, 203–208, Fall 2012
Jenny Wilson-Cohen displays her evolution simulation game at Vassar’s Darwin Days
Darwin Days at Vassar celebrates Darwin’s birthday with Jenny Wilson-Cohen demonstrating an Evolution Simulation game that she designed for her final project in her Bioinformatics Biol/CS 353 course.
Copper induces oxidative stress reponses and DNA damage in corals
Congrats to Sam Seymour ’11 and Annika O’Dea ’08 for their work analyzing the transcriptome of Montastraea franksi for our paper just out in Comparative Biochemistry and Physiology!
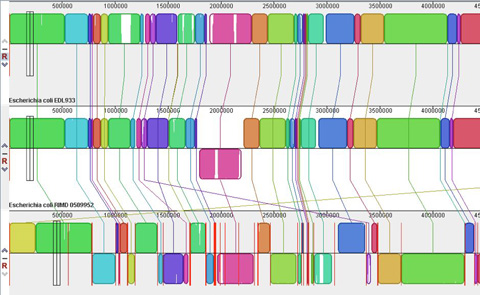
Using Comparative Genomics for Inquiry-Based Learning to Dissect Virulence
CBE-LSE 11: 81-93 Genomics and bioinformatics are topics of increasing interest in undergraduate biological science
curricula. Many existing exercises focus on gene annotation and analysis of a single genome. In
this paper, we present two educational modules designed to enable students to learn and apply
fundamental concepts in comparative genomics using examples related to bacterial pathogenesis.
David J. Baumler, Lois M. Banta, Kai F. Hung, Jodi A. Schwarz*, Eric L. Cabot, Jeremy D. Glasner, and Nicole T. Perna
CBE—Life Sciences Education, Vol. 11, 81–93, Spring 2012
Danielle Twum VC’12 investigates the role of anti-oxidants on heat-stressed anemones.
For her senior thesis project, Danielle Twum (Biology Major, 2012) examines the effects of chalcones on stress-induced bleaching of symbiotic sea anemones. Danielle is now a PhD student in Immunology at the Roswell Cancer Institute.
Genomics Field Trips to Cold Spring Harbor Labs
BIOL 244 Genomics students touring Cold Spring Harbor Labs to learn about bioinformatics and genomics research.
Generation and analysis of transcriptomic resources for a model system on the rise: the sea anemone Aiptasia pallida
Shinichi Sunagawa, Emily C Wilson, Michael Thaler, Marc L Smith, Carlo Caruso, John R Pringle, Virginia M Weis, Mónica Medina and Jodi A Schwarz*
BMC Genomics The most diverse marine ecosystems, coral reefs, depend upon a functional symbiosis between cnidarian hosts and unicellular dinoflagellate algae. The molecular mechanisms underlying the establishment, maintenance, and breakdown of the symbiotic partnership are, however, not well understood. Efforts to dissect these questions have been slow, as corals are notoriously difficult to work with. In order to expedite this field of research, we generated and analyzed a collection of expressed sequence tags (ESTs) from the sea anemone Aiptasia pallida and its dinoflagellate symbiont (Symbiodinium sp.), a system that is gaining popularity as a model to study cellular, molecular, and genomic questions related to cnidarian-dinoflagellate symbioses.