Science‘s IBI Prize was developed to showcase outstanding materials, usable in a wide range of schools and settings, for teaching introductory science courses at the college level. The materials must be designed to encourage students’ natural curiosity about how the world works, rather than to deliver facts and principles about what scientists have already discovered. With a team of scientists and educators at Carleton College and Vassar College, we developed Web-based Genomics Explorers to help guide students through the process of developing authentic research projects in genomics and bioinformatics. We were awarded the IBI Prize, accompanied by an essay published in the January 25 issue of Science.
Category Archives: Publication
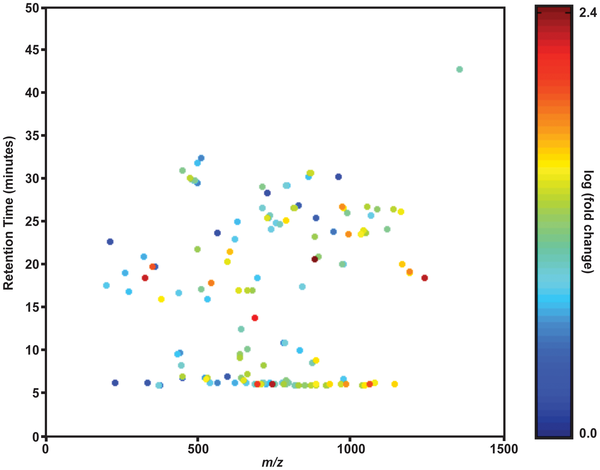
Lipid profiling of symbiosis
Symbiosis in cnidarians is accompanied by significant changes in the lipidome of the anemone host. Congrats to Josh Klein ’12, John Schmeitzel ’12, and Janice Hwang, ’12 for their authorship on this paper just out in PLoS One!
Integrating Genomics Research throughout the Undergraduate Curriculum
COVER ARTICLE in CBE-LSE: Through a multi-year, collaborative process, we developed, implemented, and peer-reviewed inquiry-based, integrated instructional units (I3Us) adaptable to a range of teaching settings, with a focus on both model and nonmodel systems. Each of the products is built on vetted design principles: 1) they have clear pedagogical objectives; 2) they are integrated with lessons taught in the lecture; 3) they are designed to integrate the learning of science content with learning about the process of science; and 4) they require student reflection and discussion. Eleven I3Us were designed and implemented as multi-week modules within the context of an existing biology course.
Lois M. Banta, Erica J. Crespi, Ross H. Nehm, Jodi A. Schwarz, Susan Singer, Cathryn A. Manduca, and workshop participants.
CBE—Life Sciences Education, Vol. 11, 203–208, Fall 2012
Copper induces oxidative stress reponses and DNA damage in corals
Congrats to Sam Seymour ’11 and Annika O’Dea ’08 for their work analyzing the transcriptome of Montastraea franksi for our paper just out in Comparative Biochemistry and Physiology!
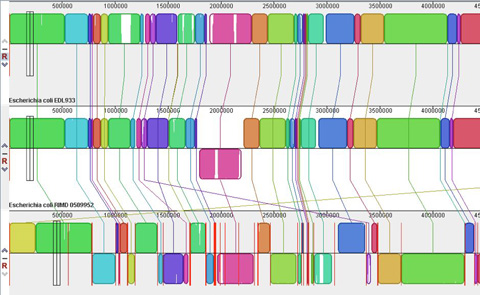
Using Comparative Genomics for Inquiry-Based Learning to Dissect Virulence
CBE-LSE 11: 81-93 Genomics and bioinformatics are topics of increasing interest in undergraduate biological science
curricula. Many existing exercises focus on gene annotation and analysis of a single genome. In
this paper, we present two educational modules designed to enable students to learn and apply
fundamental concepts in comparative genomics using examples related to bacterial pathogenesis.
David J. Baumler, Lois M. Banta, Kai F. Hung, Jodi A. Schwarz*, Eric L. Cabot, Jeremy D. Glasner, and Nicole T. Perna
CBE—Life Sciences Education, Vol. 11, 81–93, Spring 2012
Generation and analysis of transcriptomic resources for a model system on the rise: the sea anemone Aiptasia pallida
Shinichi Sunagawa, Emily C Wilson, Michael Thaler, Marc L Smith, Carlo Caruso, John R Pringle, Virginia M Weis, Mónica Medina and Jodi A Schwarz*
BMC Genomics The most diverse marine ecosystems, coral reefs, depend upon a functional symbiosis between cnidarian hosts and unicellular dinoflagellate algae. The molecular mechanisms underlying the establishment, maintenance, and breakdown of the symbiotic partnership are, however, not well understood. Efforts to dissect these questions have been slow, as corals are notoriously difficult to work with. In order to expedite this field of research, we generated and analyzed a collection of expressed sequence tags (ESTs) from the sea anemone Aiptasia pallida and its dinoflagellate symbiont (Symbiodinium sp.), a system that is gaining popularity as a model to study cellular, molecular, and genomic questions related to cnidarian-dinoflagellate symbioses.
Understanding the Intracellular Niche in Cnidarian-Dinoflagellate Symbioses: Parasites Lead the Way
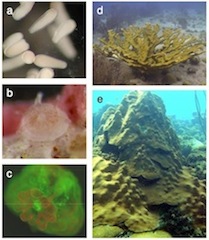
Coral life history and symbiosis: functional genomic resources for two reef building Caribbean corals
Scleractinian corals are the foundation of reef ecosystems in tropical marine environments. Their great success is due to interactions with endosymbiotic dinoflagellates (Symbiodinium spp.), with which they are obligately symbiotic. To develop a foundation for studying coral biology and coral symbiosis, we have constructed a set of cDNA libraries and generated and annotated ESTs from two species of corals, Acropora palmata and Montastraea faveolata.
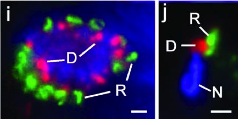
A novel rhoptry protein in Toxoplasma gondii bradyzoites and merozoites
The secretory organelles of Toxoplasma gondii orchestrate invasion of the host cell and establish the parasitophorous vacuole. Although much has been learned about the roles played by these organelles in invasion by the tachyzoite stage, little is known about the contents or functions of these organelles during bradyzoite development or pathogenesis. We identified a novel protein that localizes to the rhoptries of the bradyzoite stage, but is absent from the tachyzoite stage. This protein, BRP1, first appears in the nascent rhoptries during the first division of bradyzoite stage development.We observed secretion of BRP1 and other rhoptry proteins into the parasitophorous vacuole during bradyzoite development in vitro, but there was no evidence that this occurs in vivo. Brp1 knockout parasites did not appear to have any developmental or growth defects in vitro, and were able to establish infections in mice both as tachyzoites (via intraperitoneal injection of in vitro-derived tachyzoites) or bradyzoites (via oral gavage using cysts harvested from mouse brain). Mice infected using brain cysts from the brp1 knockout or the control strain developed similar numbers and sizes of brain cysts. Thus BRP1 does not appear to play an essential role in development of the bradyzoite stage, development of brain cysts, or oral infection of new hosts, at least in the mouse model used here. Since we also observed that BRP1 is expressed in the merozoite stages in the gut of infected cats, the coccidian phase of the life cycle may be where BRP1 plays its most important role.
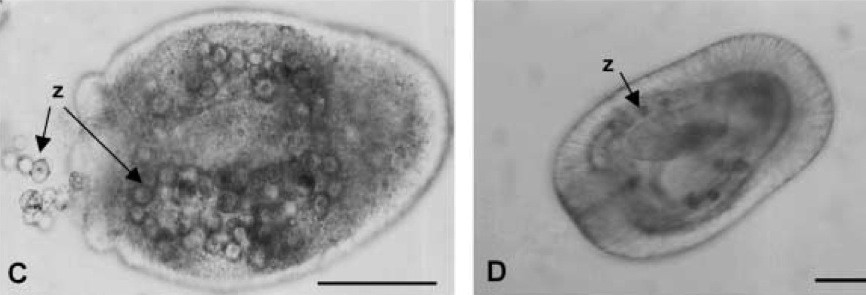
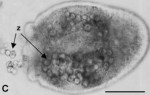
Aspects of the larval biology of the sea anemones Anthopleura elegantissima and A. artemesia
We investigated several aspects of the larval biology of the anemone Anthopleura elegantissima, which harbors algal symbionts from two different taxa, and the non-symbiotic A. artemisiu. From a 7-year study, we report variable spawning and fertilization success of A. elegantissima in the laboratory. We examined the dynamics of symbiosis onset in larvae of A . elegantissima. Zoochlorellae, freshly isolated from an adult host, were taken up and retained during the larval feeding process, as has been described previously for zooxanthellae. In addition, larvae infected with zooxanthellae remained more highly infected in high-light conditions, compared to larvae with zoochlorellae, which remained more highly infected in low-light conditions. These results parallel the differential distribution of the algal types observed in adult anemones in the field and their differential tolerances to light and temperature. We report on numerous failed attempts to induce settlement and metamorphosis of l a rva e of A . eleguntissima, using a variety of substrates and chemical inducers. We also describe a novel change in morphology of some older planulae, in which large bulges, resembling tentacles, develop around the mouth. Finally, we provide the first description of planulae of A. artemisia and report on attempts t o infect this non-symbiotic species with xooxanthellae and zoochlorellae.